Applications
Wastewater Epidemiology
Wastewater-based epidemiology (WBE) aims at measuring pathogens in wastewater at population scale.
This approach has proven to enable advanced warning ahead of clinical testing or hospitalizations, for public agencies and communities to better prepare for and prevent the spread of infectious diseases.
Hyperplex PCR™ is currently being used to monitor the spread of COVID variants in Sweden together with the Swedish Environmental Epidemiology Centre (SEEC) and is now available to be deployed world-wide.

Sites currently monitored in Sweden with hpPCR
18
Data points analyzed since Q4 2022
15 800+
Multiplex currently in panel
18
Why Hyperplex PCR for pandemic preparedness?

With multiplexing unleashed, Hyperplex PCR enables shorter time to action by enabling instant set-up and in real-time updating of panels with unparallaled specificity and sensitivity for routine monitoring and better pandemic preparedness systems.
What we measure
SARS-CoV-2
variant panel
18 mutations to discriminate 15 targets
PMMoV
XBB
XBB.1.5
EG.1
N3
CH.1.1
XBB.1.9
XBB.1.9.1/2
BA.2.75
EG.5
XBB.1.16
BA.2.86
BQ.1.1
EG.5.1
XBB.1.16.1
Other markers available or of importance to public health
Any target can be mix-and-matched into a new panel or on top of an existing panel
Influenza A
Influenza B
AMR sul1
Norovirus GII
HCoV NL63
Adenovirus 40/41
Influenza A H1
AMR mecA
AMR mefA
RSV A
HCoV 223E
Hepatitis A
Influenza A H3
AMR oxa-48
AMR 16s rRNA
RSV B
HCoV OC43
Rotavirus
Influenza A H5 (avian)
AMR tetA
Norovirus GI
Mpox virus
HCoV HKU1
SARS-CoV-2 variants surveillance data
Together with the Swedish Environmental Epidemiology Center (SEEC), SARS-CoV-2 variants in Sweden are being monitored using Hyperplex PCR.
Up to 22 sites have been monitored with a 18-plex panel enabling monitoring unparallalled by other methods such as qPCR, dPCR and NGS.
The panel as been adapted to keep it up-to-date with relevant variants of concern by removing exting variants and swiftly add new mutations within 2 weeks from order.
Read about some of the conclusions reached in our application note.
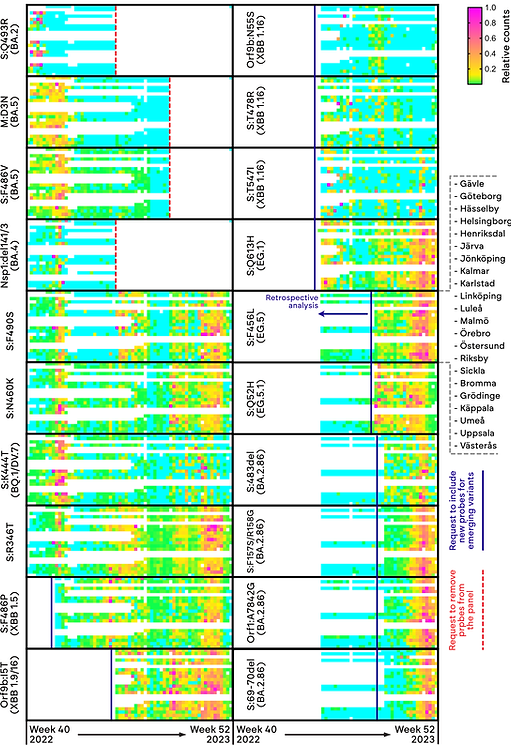
Figure 1. Heatmap summary of SARS-CoV-2 VOC surveillance data generated with Hyperplex PCR over one year with an adaptable 18-plex panel for up to 22 sites in Sweden. Heatmap colors represent as following; blue (no signal detected), from green (low concentration) to pink (high concentration) of marker detected.
NGS-grade quantification of mutation frequency

Figure 2. A, B: Mutation frequency measured in wastewater with NGS (filled symbols) and hpPCR (open symbols). C: Linearity of mutation frequency measurements using hpPCR in a 10-plex target system (spiked targets). D: hpPCR vs NGS mutation frequency in WW samples.
4+ weeks earlier detection of emerging variants

Figure 3. Weekly measurement of S:F486P, characteristic of XBB 1.5 using hpPCR and NGS (Ion torrent). 8 wastewater collection sites were measured at each time point. Both measurements were performed on the same extracted nucleic acid sample.
Inhibitors problematic for wastewater samples

Figure 4. Wastewater matrix contains inhibitors affecting the amplification in both qPCR and dPCR measurements. While dPCR may handle this better than qPCR, it is still subject to variance in analysis which has effect on the accuracy of analysis.
Negligible inhibitor effect with hpPCR

Figure 5. Intensity of N3 gene marker obtained after hpPCR on DNAse/RNAse-free water (pink) and 29 different wastewater samples (green). Signals from the control (pink) and wastewater samples show no significant difference, and both samples are well above the limit of detection (LoD) for signal detection of RCPs, making in particular mutation frequency analysis virtually not effected by inhibitors at all.
AMR analysis of low and high abundance targets
In this project, Hyperplex PCR was used for absolute quantification of AMR markers in wastewater samples, showcasing the read-out of targets as low as less than 10 copies/uL, combined with others as high as 105 copies/uL.
Expand the panel with more AMR markers and other pathogens of your choice while retaining absolute quantification.
